
CoderData is a data assembly pipeline that pulls from original data sources of drug sensitivity and omics datasets and assembles them so they can be integrated into a python package for AI/ML integration.
CoderData is indeed a work in progress. If you have specific requests or bugs, please file an issue on our GitHub page and we will begin a conversation about details and how to fix the issue. If you would like to create a new feature to address the issue, you are welcome to fork the repository and create a pull request to discuss it in more detail. These will be triaged by the CoderData team as they are received.
The rest of this document is focused on how to contribute to and augment CoderData, either for use by the community or your own purposes.
To build your own internal Coderdata dataset, or to augment it, it is important to understand how the package is built.
The build process is managed in the build
directory
primarily by the build_all.py script. The
build_dataset.py script is used for testing the development of each dataset in CoderData.
Because our sample and drug identifiers are unique, we must
finish the generation of one dataset before we move to the next. This
process is depicted below.
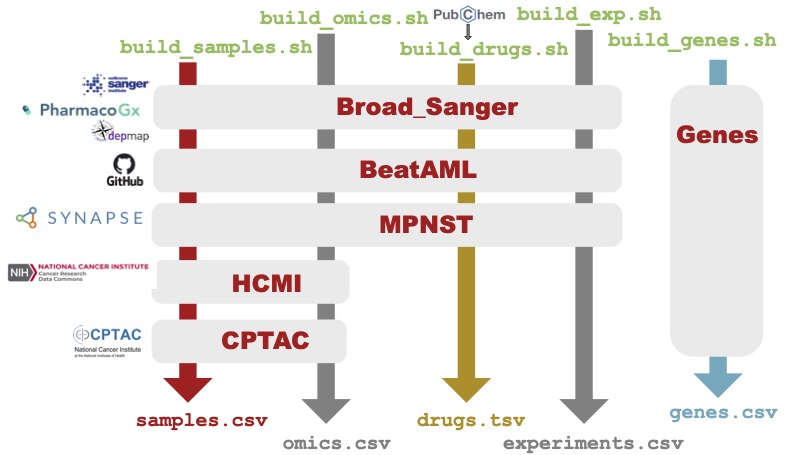
The build process is slow, partially due to our querying of PubChem, and also because of our extensive curve fitting. However, it can be run locally so that you can still leverage the Python package functionality with your own datasets.
If you want to add a new dataset, you must create a docker image that
contains all the scripts to pull the data and reformat it into our
LinkML Schema. Once complete, you can modify build_dataset.py to
call your Docker image and associated scripts. More details are below.
To add your own data, you must add a Docker image with the following constraints:
Dockerfile.[dataset_name] and reside in the
/build/docker directorybuild_omics.sh, build_samples.sh,
build_drugs.sh, build_exp.sh , and if needed, a
build_misc.sh. These will all be called directly by
build_dataset.py.Files are generated in the following order as described below.
The first step of any dataset build is to create a unique set of
sample identifies and store them in a [dataset_name]_samples.csv
file. We recommend following these steps:
build_samples.sh script that calls your script with an
existing sample file as the first argument.build_samples.sh script with a test sample
file.The overall omics generation process is the same as the samples, with a few caveats.
build_omics.sh script that calls your script with the
genes.csv file as the first argument and [dataset_name]_samples.csv file as second
argument.build_omics.sh script with your sample file and test genes
file.The precise data files have varying standards, as described below:
The resulting files are then stored as [dataset_name]_[datatype].csv.
The drug generation process can be slow depending on how many drugs require querying from PubChem. However, with the use of an existing drug data file, it’s possible to shorten this process.
published_auc as the drug_response_metric value in the table.build_drugs.sh script that takes as its first argument
an existing drug file and calls the script created in step 1
above. Once the drugs for a dataset are retrieved, we have a second utility
script that builds the drug descriptor table. Add this to the
shell script to generate the drug descriptor file.build_drugs.sh script with the test drugs
file.The resulting files should be [dataset_name]_drugs.tsv and
[dataset_name]_drug_descriptors.tsv.
The experiment file maps the sample information to the drugs of interest with various drug response metrics. The experiment data varies based on the type of system:
Otherwise the steps for building an experiment file are similar:
build_exp.sh script that takes as its first argument
the samples file and the second argument the drug file.build_exp.sh script with the drug and samples files.All scripts described above go into a single directory with the name of the dataset under the build directory, with instructions to add everything in the docker directory. Make sure to include any requirements for building in the folder and docker image as well.
Once the Dockerfile builds and runs, you can modify the
build_dataset.py script so that it runs and validates.
Check out examples! We have numerous Docker files in our Dockerfile directory, and multiple datasets in our build directory.
Your contributions are essential to the growth and improvement of CoderData. We look forward to collaborating with you!