Contribution¶
How to Contribute¶
Contribute to CoderData¶
CoderData is a data assembly pipeline that pulls from original data sources of drug sensitivity and omics datasets and assembles them so they can be integrated into a python package for AI/ML integration.
CoderData is indeed a work in progress. If you have specific requests or bugs, please file an issue on our GitHub page and we will begin a conversation about details and how to fix the issue. If you would like to create a new feature to address the issue, you are welcome to fork the repository and create a pull request to discuss it in more detail. These will be triaged by the CoderData team as they are received.
The rest of this document is focused on how to contribute to and augment CoderData, either for use by the community or your own purposes.
CoderData build process¶
To build your own internal Coderdata dataset, or to augment it, it is important to understand how the package is built.
The build process is managed in the build
directory
primarily by the build_all.py script. The
build_dataset.py script is used for testing the development of each dataset in CoderData.
Because our sample and drug identifiers are unique, we must
finish the generation of one dataset before we move to the next. This
process is depicted below.
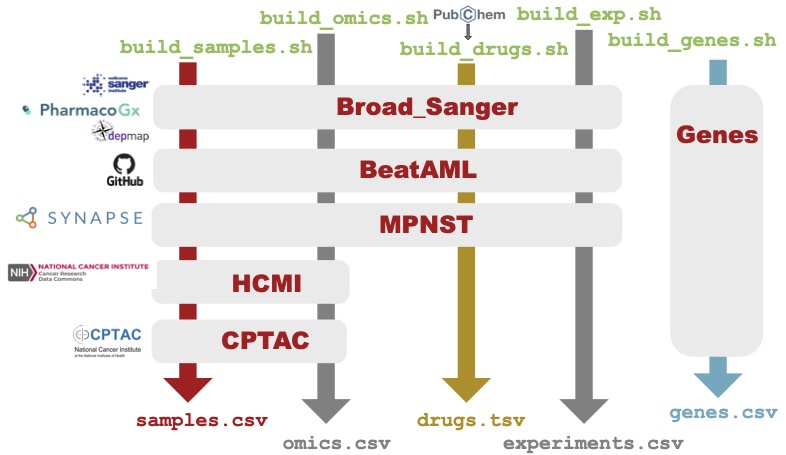
The build process is slow, partially due to our querying of PubChem, and also because of our extensive curve fitting. However, it can be run locally so that you can still leverage the Python package functionality with your own datasets.
If you want to add a new dataset, you must create a docker image that
contains all the scripts to pull the data and reformat it into our
LinkML Schema. Once complete, you can modify build_dataset.py to
call your Docker image and associated scripts. More details are below.
Adding your own dataset¶
To add your own data, you must add a Docker image with the following constraints:
Be named
Dockerfile.[dataset_name]and reside in the/build/dockerdirectoryPossess scripts called
build_omics.sh,build_samples.sh,build_drugs.sh,build_exp.sh, and if needed, abuild_misc.sh. These will all be called directly bybuild_dataset.py.Create tables that mirror the schema described by the LinkML YAML file.
Files are generated in the following order as described below.
Sample generation¶
The first step of any dataset build is to create a unique set of
sample identifies and store them in a [dataset_name]_samples.csv
file. We recommend following these steps:
Build a python script that pulls the sample identifier information from a stable repository and generates Improve identifiers for each sample while also ensuring that no sample identifiers are clashing with prior samples. Examples can be found here and here. If you are using the Genomic Data Commons, you can leverage our existing scripts here.
Create a
build_samples.shscript that calls your script with an existing sample file as the first argument.Test the
build_samples.shscript with a test sample file.Validate the file with the linkML validation tool and our schema file.
Omics data generation¶
The overall omics generation process is the same as the samples, with a few caveats.
Build a python script that maps the omics data and gene data to the standardized identifiers and aligns them to the schema. pulls the sample identifier information from a stable repository and generates Improve identifiers for each sample while also ensuring that no sample identifiers are clashing with prior samples. Examples can be found here and here. If you are using the Genomic Data Commons, you can leverage our existing scripts here. For each type of omics data (see below), a single file is created.It might take more than one script, but you can combine those in step 2.
Create a
build_omics.shscript that calls your script with thegenes.csvfile as the first argument and[dataset_name]_samples.csvfile as second argument.Test the
build_omics.shscript with your sample file and test genes file.Validate the files generated with the linkML validation tool and our schema file.
The precise data files have varying standards, as described below:
Mutation data: In addition to matching gene identifiers each gene mutation should be mapped to a specific schema of variations. The list of allowed variations can be found in our linkML file.
Transcriptomic data: Transcript data is mapped to the same gene identifiers and samples but is convered to transcripts per million, or TPM.
Copy number data: Copy number is assumed to be a value representing the number of copies of that gene in a particular sample. A value of 2 is assumed to be diploid.
Proteomic data: Proteomic measurements are generally log ratio values of the abundance measurements normalized to an internal control.
The resulting files are then stored as [dataset_name]_[datatype].csv.
Drug data generation¶
The drug generation process can be slow depending on how many drugs require querying from PubChem. However, with the use of an existing drug data file, it’s possible to shorten this process.
Build a python script that maps the drug information to SMILES String and IMPROVE identifier. All drugs are given an Improve identifier based on the canonical SMILES string to ensure that each drug has a unique structure to be used in the modeling process. To standardize this we encourage using our standard drug lookup script that retrieves drug structure and information by name or identifier. This file of NCI60 drugs is our most comprehensive script as it pulls over 50k drugs 1a. In cases where the dose and response values are not available, you can use the published AUC values instead, and use the
published_aucas thedrug_response_metricvalue in the table.Create a
build_drugs.shscript that takes as its first argument an existing drug file and calls the script created in step 1 above. Once the drugs for a dataset are retrieved, we have a second utility script that builds the drug descriptor table. Add this to the shell script to generate the drug descriptor file.Test the
build_drugs.shscript with the test drugs file.Validate the files generated with the linkML validation tool and our schema file.
The resulting files should be [dataset_name]_drugs.tsv and
[dataset_name]_drug_descriptors.tsv.
Experiment data generation¶
The experiment file maps the sample information to the drugs of interest with various drug response metrics. The experiment data varies based on the type of system:
Cell line and organoid data use the drug curve fitting tool that maps doses of drugs (in Moles) to drug response measurements (in percent) to a variety of curve fitting metrics described in our schema file.
Patient derived xenografts require an alternate script that creates PDX-speciic metrics.
Otherwise the steps for building an experiment file are similar:
Build a python script that maps the drug information and sample information to the DOSE and GROWTH values, then calls the appropriate curve fitting tool described above.
Create a
build_exp.shscript that takes as its first argument the samples file and the second argument the drug file.Test the
build_exp.shscript with the drug and samples files.Validate the files generated with the linkML validation tool and our schema file.
Dockerize and test¶
All scripts described above go into a single directory with the name of the dataset under the build directory, with instructions to add everything in the docker directory. Make sure to include any requirements for building in the folder and docker image as well.
Once the Dockerfile builds and runs, you can modify the
build_dataset.py script so that it runs and validates.
Check out examples! We have numerous Docker files in our Dockerfile directory, and multiple datasets in our build directory.
Your contributions are essential to the growth and improvement of CoderData. We look forward to collaborating with you!
Guide to Adding Code to CoderData¶
Introduction¶
This guide outlines the steps for contributors looking to add new code functionalities to CoderData, including automation scripts, Docker integration, and continuous integration processes.
1. Automate Credentials and Data Pulling¶
Script Development: Write a script to automate credential management and data pulling, ensuring secure handling of API keys or authentication tokens.
Data Formatting: The script should reformat the pulled data to fit CoderData’s existing schema.
Environment Variables: Store sensitive information such as API keys in environment variables for security.
2. Writing Tests¶
Unit Tests: Create unit tests for each function in your script.
Data Tests: Implement data tests to verify your data is accessing APIs correctly, properly formatted, and matches the schema.
3. Dockerization¶
Dockerfile Creation: Write a Dockerfile to containerize your script, specifying dependencies, environment setup, and entry points.
Local Testing: Test the Docker container locally to confirm correct functionality.
4. GitHub Actions for Automation¶
Workflow Setup: Design a GitHub Actions workflow to automate the script execution. You may directly update our version, or create a seperate workflow and we could join it to ours. This should include:
Trigger mechanisms (schedule or events).
Secret management for credentials.
Error logging and handling.
Seperate steps for samples file generation and the rest of the data generation.
5. Updating Documentation¶
Documentation Revision: Update the project documentation to reflect your new script’s purpose and usage.
Usage Examples: Provide clear examples and usage instructions.
6. Continuous Integration¶
CI Integration: Integrate your script into the existing Continuous Integration pipeline.
CI Testing: Ensure your script is included in the CI testing process to prevent future breakages.
7. Pull Request and Code Review¶
Create a Pull Request: Submit your changes via a pull request to the main CoderData repository.
8. Align with Repository Standards¶
Review Project Guidelines: Familiarize yourself with the CoderData project’s standards and practices to align your contribution accordingly.
Conclusion¶
Contributors are encouraged to follow these guidelines closely for effective integration of new code into the CoderData project. We appreciate your contributions and look forward to your innovative enhancements to the project.
Your contributions are essential to the growth and improvement of CoderData. We look forward to collaborating with you!
